Cellular mix-ups can set the stage for kidney disease
May 5, 2025
Our kidneys perform many important jobs.
They filter our blood to remove waste, regulate blood pressure by eliminating excess fluid and ensure our electrolytes are balanced, which helps keep systems throughout the body up and running.
All these vital functions depend on our kidneys having the right mix of cells. Too much of one cell type or too little of another can disrupt that balance and set the stage for disease.
“Our genes contain the blueprints for building healthy kidneys but sometimes, genes can be damaged or the instructions in genes can be misread,” said VAI’s Dr. Hong Wen. “When this happens, the kidneys may not develop properly. We want to understand the factors that affect kidney development so we can better address — and hopefully even fix — these problems.”
Some of these issues are linked to epigenetics, a complex set of mechanisms that determines how and when the instructions in our genes are used without changing the gene itself. Unlike genetic changes, epigenetic changes are reversible, making them an increasingly popular target for potential new treatments.
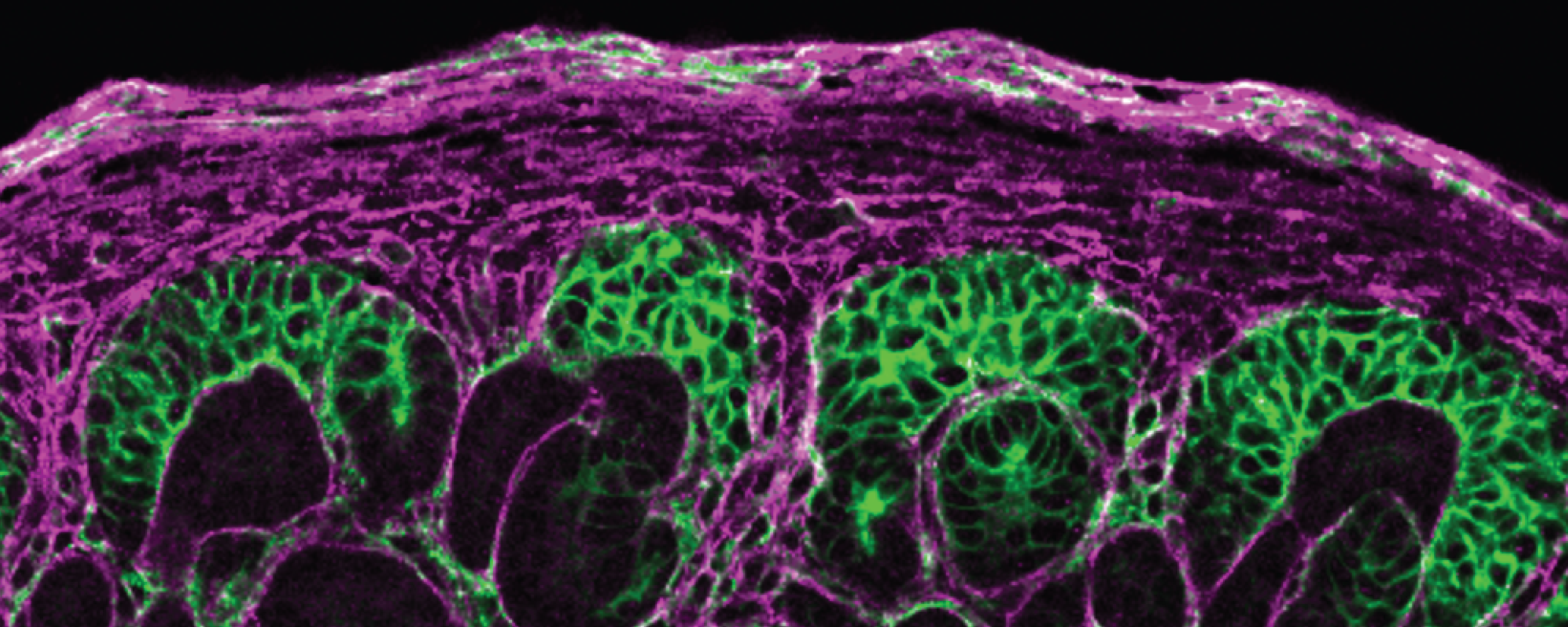
A recent study by Wen and her collaborators revealed how an epigenetic regulator called ENL influences cell fate and kidney development. The findings were published in the journal Nature Communications.
Cell fate is the process by which a cell becomes committed to a particular function or role in the body. For instance, some cells provide supportive tissues (stromal cells), while others form the functional units of the kidney (nephron cells).
This decision, however, is not up to the cells themselves.
“As an epigenetic regulator, ENL is in charge of deciding what role each cell is going to take on,” Wen explained. “When changes to ENL occur, it can have devastating consequences for the kidney over time.”
Certain variations in ENL can contribute to Wilms tumor, a type of kidney cancer that primarily affects children. Wen and her team investigate how these changes can lead normal kidney cells down the wrong path.
Using a cutting-edge method called spatial transcriptomics, they found that changes to ENL can cause the cells that form kidneys to become mixed up, impairing kidney development. Spatial transcriptomics is a technique that helps scientists to map the activity of genes within tissues to better identify cell type and location.
The findings offer new insights that pave the way toward identifying potential ways to treat kidney disease.
“Better understanding these important early processes may provide new opportunities for fixing ENL and treating kidney disease,” Wen said. “We may even be able to create specifically designed tools that can target and break down harmful proteins and improve disease treatments.”
Funding Acknowledgement
Research reported in this publication was supported by Van Andel Institute; the National Cancer Institute of the National Institutes of Health under award nos. R01CA260666 (Wen and Jin), R01CA255506 (Wen) and R01CA268440 (Shi). Wen is a Career Development Award Scholar of the Leukemia & Lymphoma Society (award no. 1387-23). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or other funding organizations.

